

by Sung-Hui Yi, Scientist Protein Biochemistry & Luisa Gronenberg, Head of R&D for Functional Ingredients & Strain Engineering
Scent, flavor, color, and other properties are what make our favorite food and personal care products particularly special. Increasingly, these ingredients come from exploitative processes. At Insempra we build microbial strains that can convert renewable feedstocks to natural ingredients by precision fermentation – a natural process that is non-exploitative.
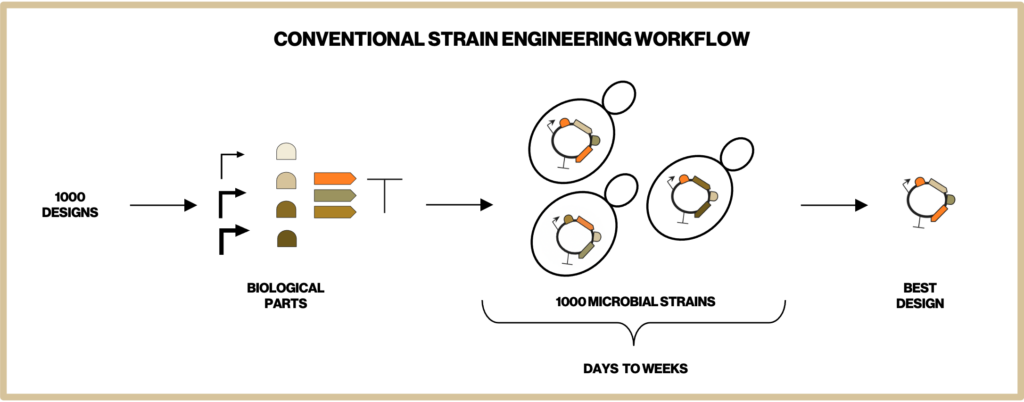
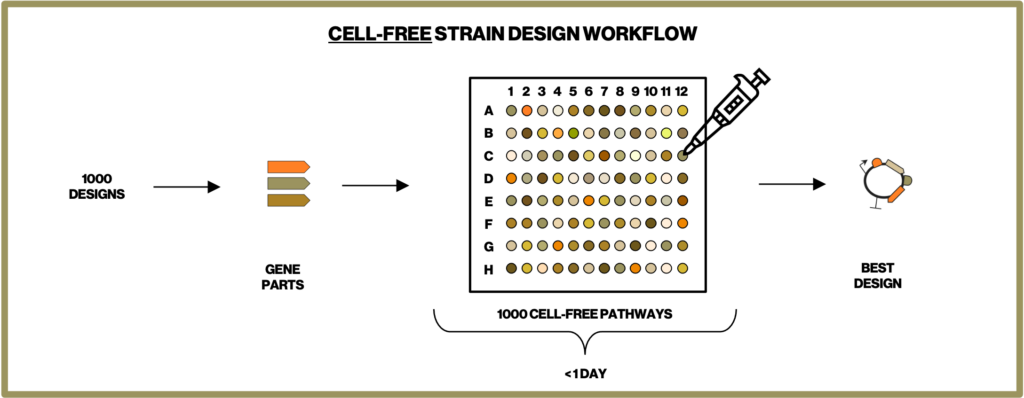
To make the most efficient microbes for producing our natural ingredients we have to screen through thousands of metabolic designs. Building each of these designed microbial strains is one of the most time-intensive steps in our development process. Therefore, at Insempra, we have built a cell-free platform to test metabolic designs quickly and accelerate time to market.
We have set up our cell-free system as a fast-screening platform together with our scientific advisor Prof. Michael Jewett from Northwestern University. Jewett is a pioneer in the area of cell-free metabolic engineering, building metabolic pathways in cell lysate as a way to directly access and control the many “knobs” of metabolism such as protein variants, protein levels and concentration of cofactors. In recent years, his group has applied cell-free systems to the synthesis of numerous sustainable bioproducts, like styrene (Grubbe, Metabolic Engineering, 2020), butanol (Karim, Nature Chemical Biology, 2020), limonene (Dudley, Metabolic Engineering, 2021), acetone (Liew, Nature Biotechnology, 2022), and hexanol (Vogeli, Nature Communications, 2022).
One of the major ways we apply the cell-free systems is by Cell-Free Protein Synthesis (CFPS). In CFPS, the transcription and translation machinery are reconstructed in cell-free reactions. By introducing DNA bearing one or multiple genes of interest, we are able to synthesize target proteins within a few hours and characterize their functions immediately. As shown in Figure 1 this eliminates the time-consuming steps in conventional strain construction of cloning of a gene into a plasmid, transformation of the plasmid into a microbial cell, culturing and harvesting the cells (and potentially purifying the protein) before assaying the protein function. This allows us to compare the efficiency of single or combinations of enzymes and narrows down the number of designs before we construct complex pathways in cells. Our cell-free process decreases the time required to achieve desired process objectives.
Sources:
Vogeli B, Schulz L, Garg S, Tarasava K, Clomburg JM, Lee SH, Gonnot A, Moully EH, Kimmel BR, Mrksich M, Karim AS, Gonzalez R, Kopke M, Jewett MC. (2022) “Cell-free prototyping enables implementation of optimized reverse b-oxidation pathways in heterotrophic and autotrophic bacteria” Nature Communications, 13: 3058
Grubbe WS, Rasor BJ, Krüger A, Jewett MC, Karim AS (2020) “Cell-free styrene biosynthesis at high titers” Metabolic Engineering 61: 89-95
Dudley QM, Karim AS, Nash CJ, Jewett MC (2020) “In vitro prototyping of limonene biosynthesis using cell-free protein synthesis” Metabolic Engineering 61: 251-260
Liew, F.,† Nogle, R.,† Abdalla, T.,† Rasor, B.J., Canter, C., Jensen,R.O., Wang, L., Strutz, J., Chirania, P., Tissera, S.D., Mueller, A.P., Ruan, Z., Gao, A., Tran, L., Bromley, J.C., Daniell, J., Conrado, R., Tschaplinski, T.J., Giannone, R.J., Hettich, R.L., Karim, A.S., Simpson, S.D., Brown, S.D., Leang, C.,* Jewett, M.C.,* and Köpke, M.* 2022. Carbon-negative production of acetone and isopropanol by gas fermentation at industrial pilot scale.
Karim, A.S., Dudley, Q.M., Juminaga, A., Yuan, Y., Crowe, S.A., Heggestad, J.T., Garg, S., Abdalla, T., Grubbe, W.,% Rasor, B., Coar, D., Torculas, M., Krein, M., Liew, F., Quattlebaum, A., Jensen, R.O., Stuart, J., Simpson, S.D., Köpke, M., and Jewett, M.C.* 2020. In vitro prototyping and rapid optimization of biosynthetic enzymes for cellular design. Nature Chemical Biology. 16: 912-919.
Photo by Louis Reed on Unsplash